In 2014, I was awarded funding to perform a research project as a Marie Curie fellow at the University of Edinburgh at the institute for condensed matter and complex systems in the groups physics of living matter and soft matter physics.
Bacteria as living colloids
Bacteria are essentially living colloids, have micrometer dimensions, and are also subject to Brownian motion. However, bacteria are active in the sense they can put internal energy into work to propel themselves and swim inside a fluid. In addition, they can rapidly multiply through division and under some conditions cause problems to humans and other animals, such as infections. Besides their ubiquity in the real world, bacteria such as Escherichia coli (E. coli) are used as model active colloids in the laboratory. Their swimming speed and physical properties are well-characterised and can be tuned and controlled.
Computer simulations of bacteria and colloidal spheres
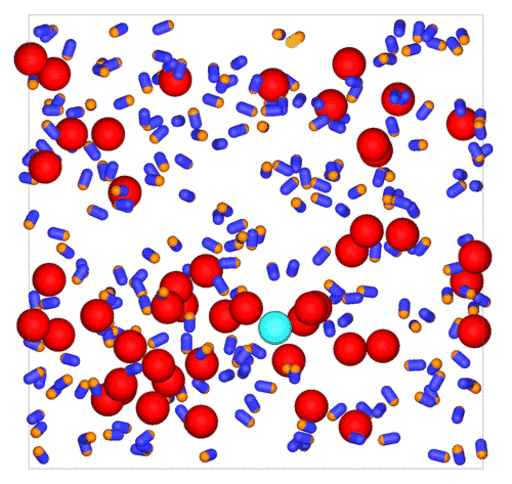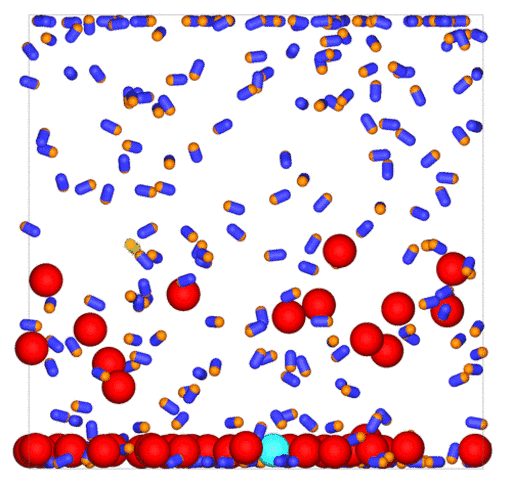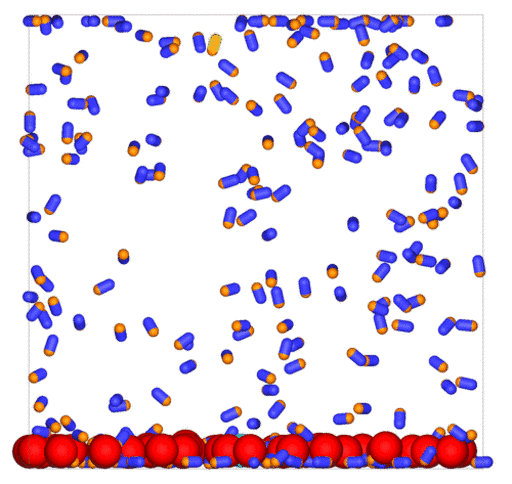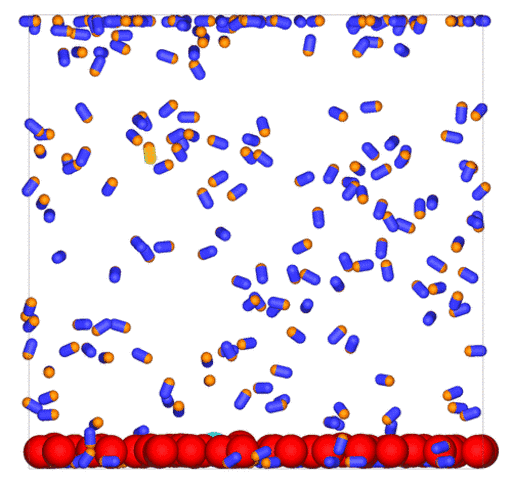
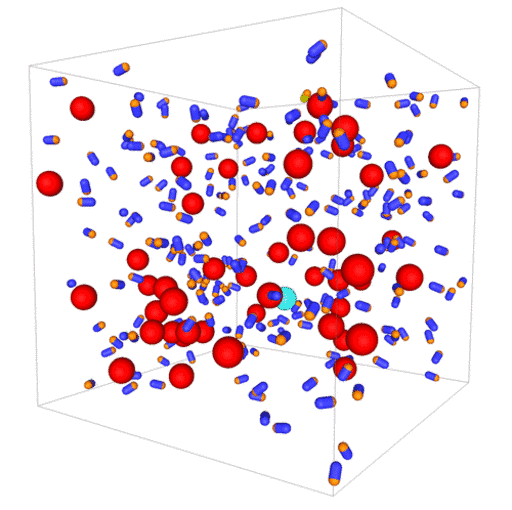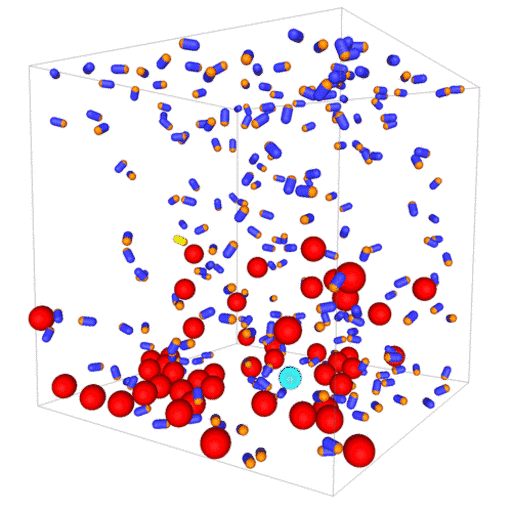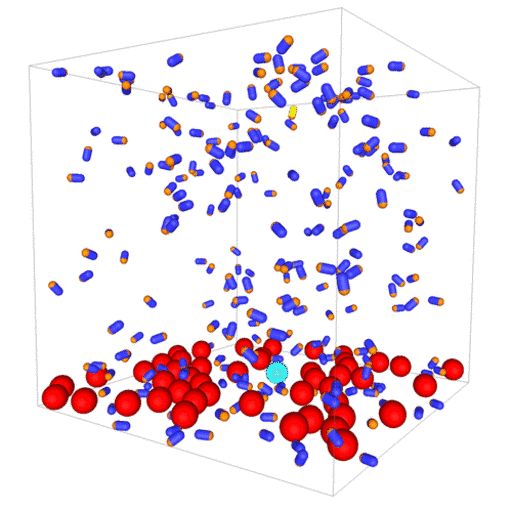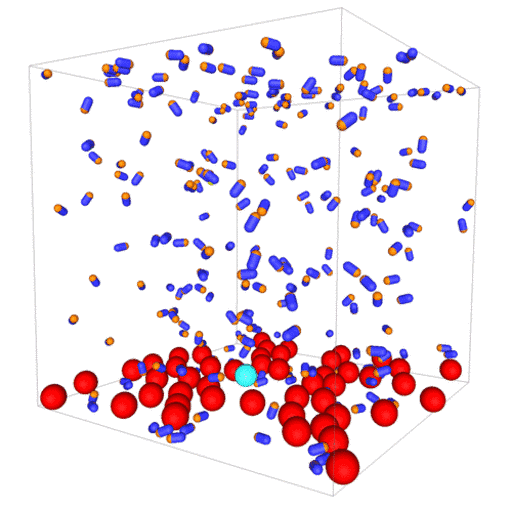
Passive colloids in a dispersion undergo Brownian motion. However, when spherical colloids are mixed with bacteria, the bacteria can `kick’ them around, which has a strong effect on their overall dynamics. Computer simulations can quantify these dynamics to explain the experiments. The videos below show a side and perspective view of a computer simulation. The swimming motion is very much simplified, and many features are left out. The colloids move diffusively and the bacteria move in a run-an-tumble motion fashion. Bacteria and spherical colloids interact with each other via a short-ranged repulsive potential in a similar way to what is described here.
Bacteria on surfaces
Billions of years of evolution have optimised bacteria to exploit a wide range of ecological niches on earth. From deep see troughs to rocks, plant roots, animal guts, surfaces make up a large part of their habitat. In time, bacteria have become masters of using and colonising these surfaces as well as man-made surfaces to form biofilms, that protect them from physical and chemical assaults. In particular, the ability of microbes to form drug-resistant biofilms on medical equipment, implant organs, food packaging, and water distribution systems presents major healthcare and economic challenges. Strategies to deal with these challenges require a fundamental understanding of bacteria-surface interactions.
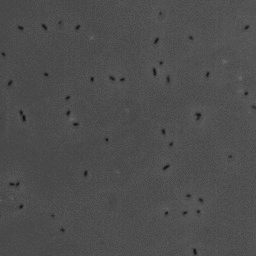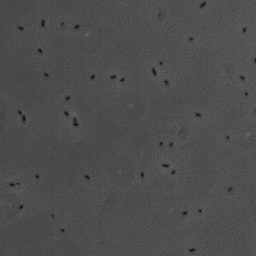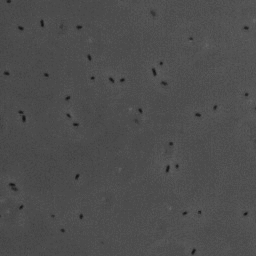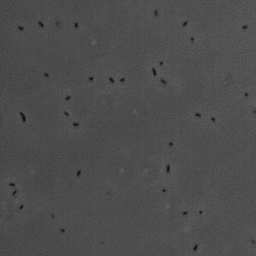
In this project, we have investigated adhesion of bacteria to surfaces, and specifically how, and under which conditions individual cells adhere. The video below shows rod-shaped E. coli bacteria on a glass surface. Some bacteria are moving, whereas others are adhering to the surface. The video was recorded using phase contrast microscopy. The bacteria are a few micrometer in length, and have a diameter that is a bit less than a micrometer.
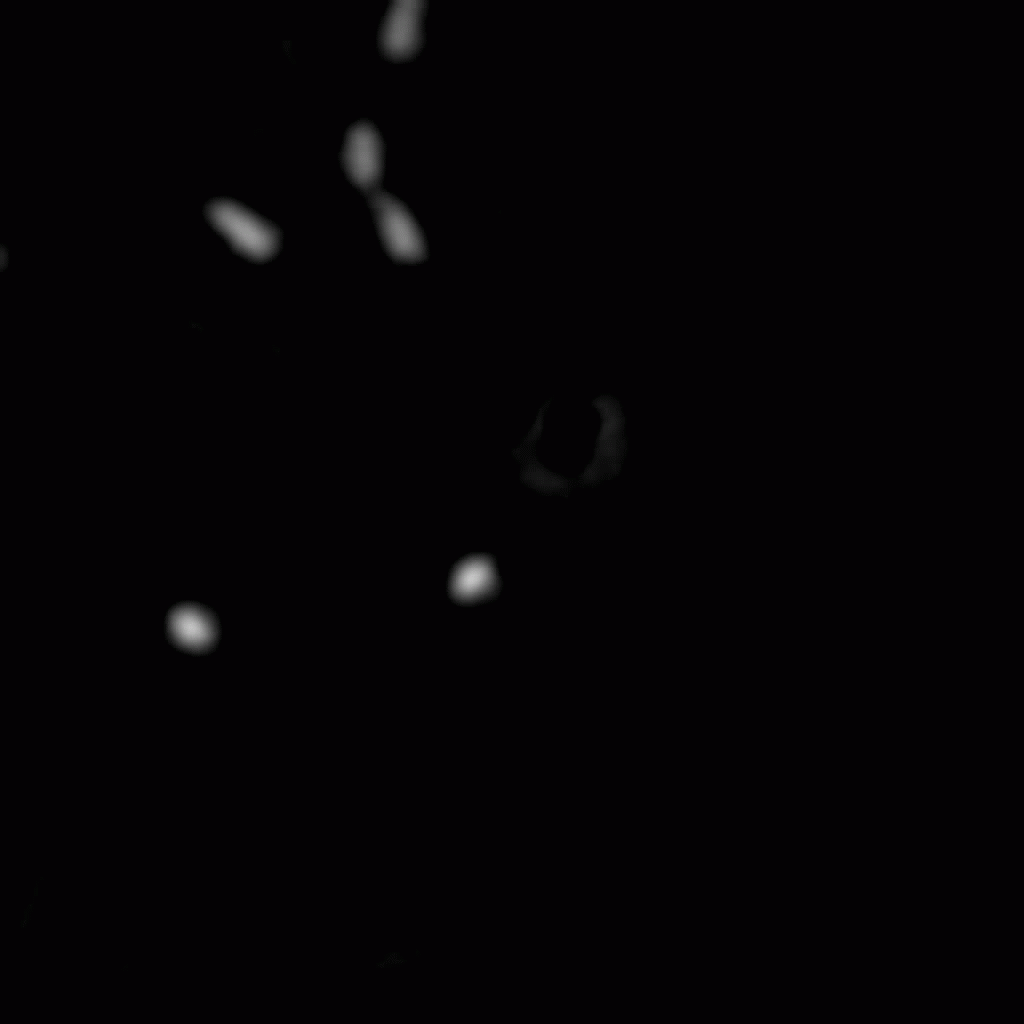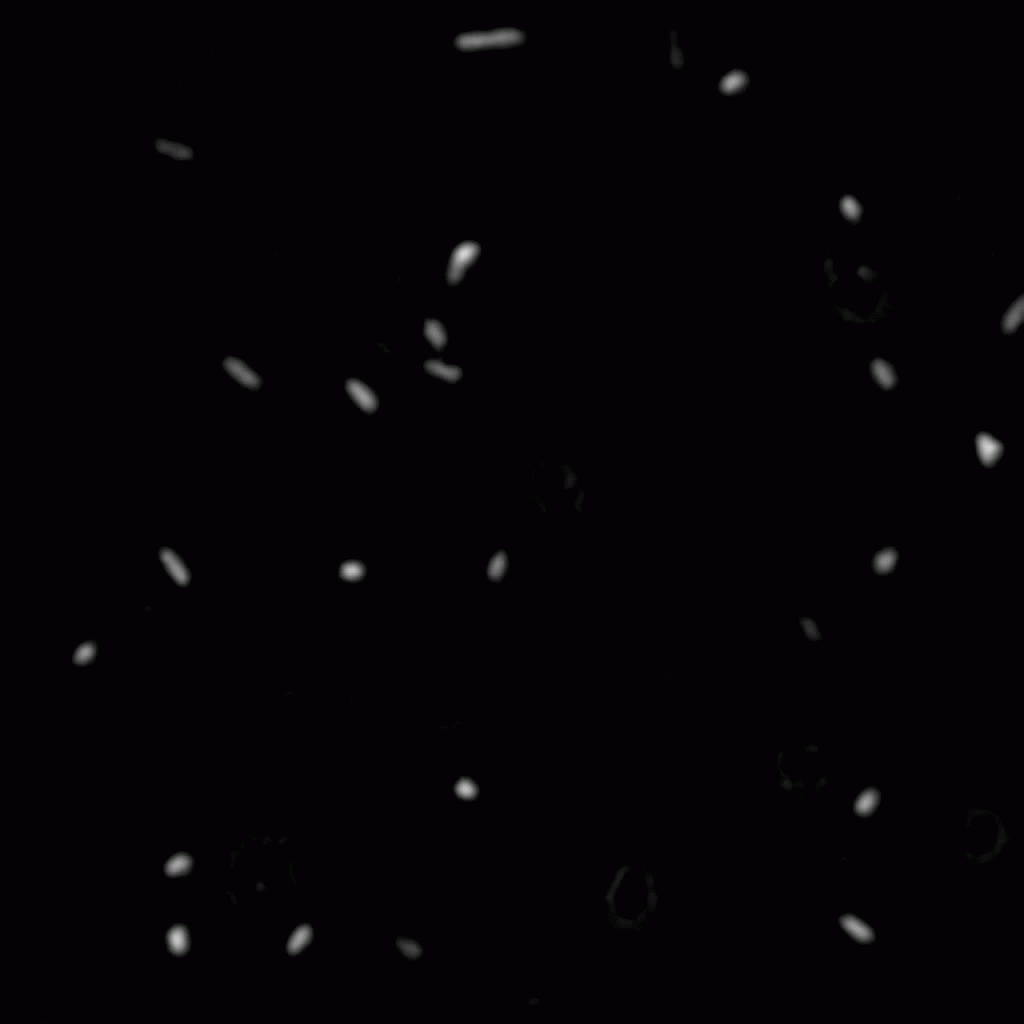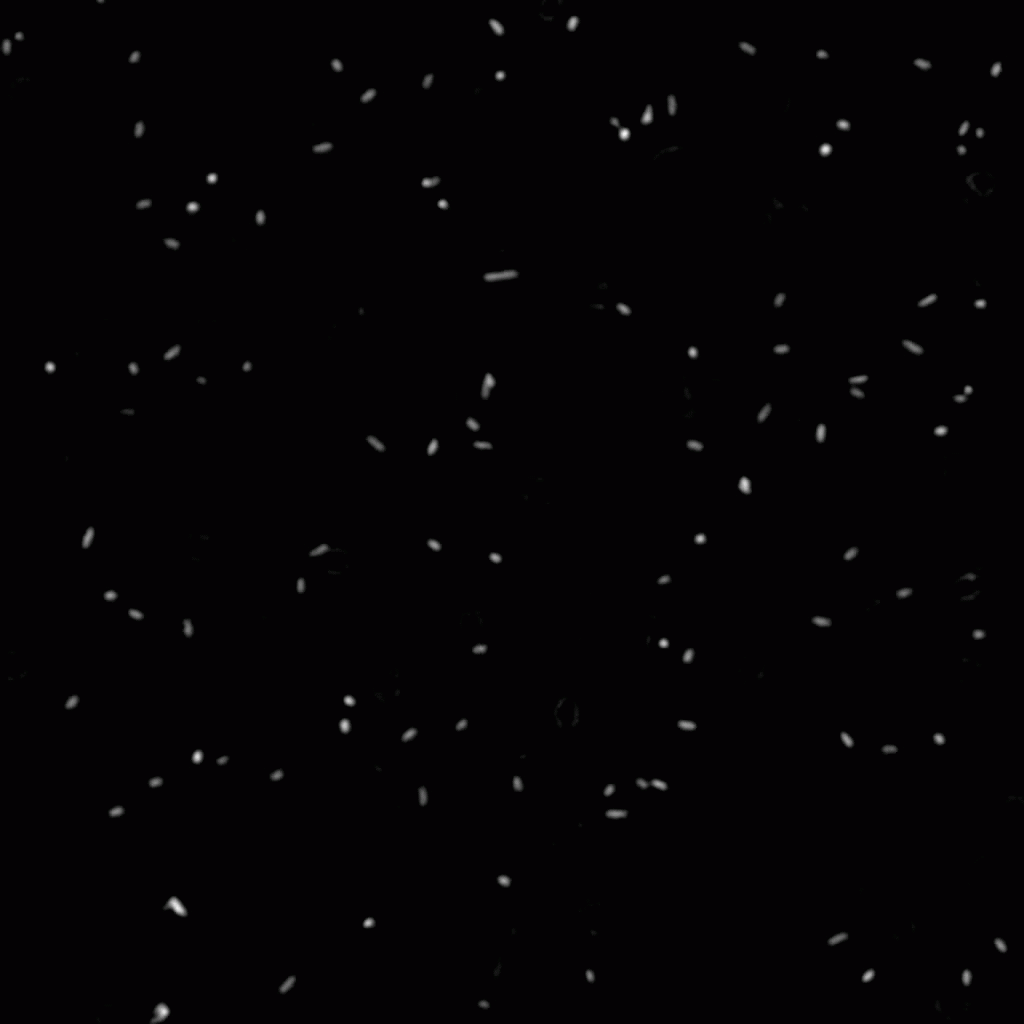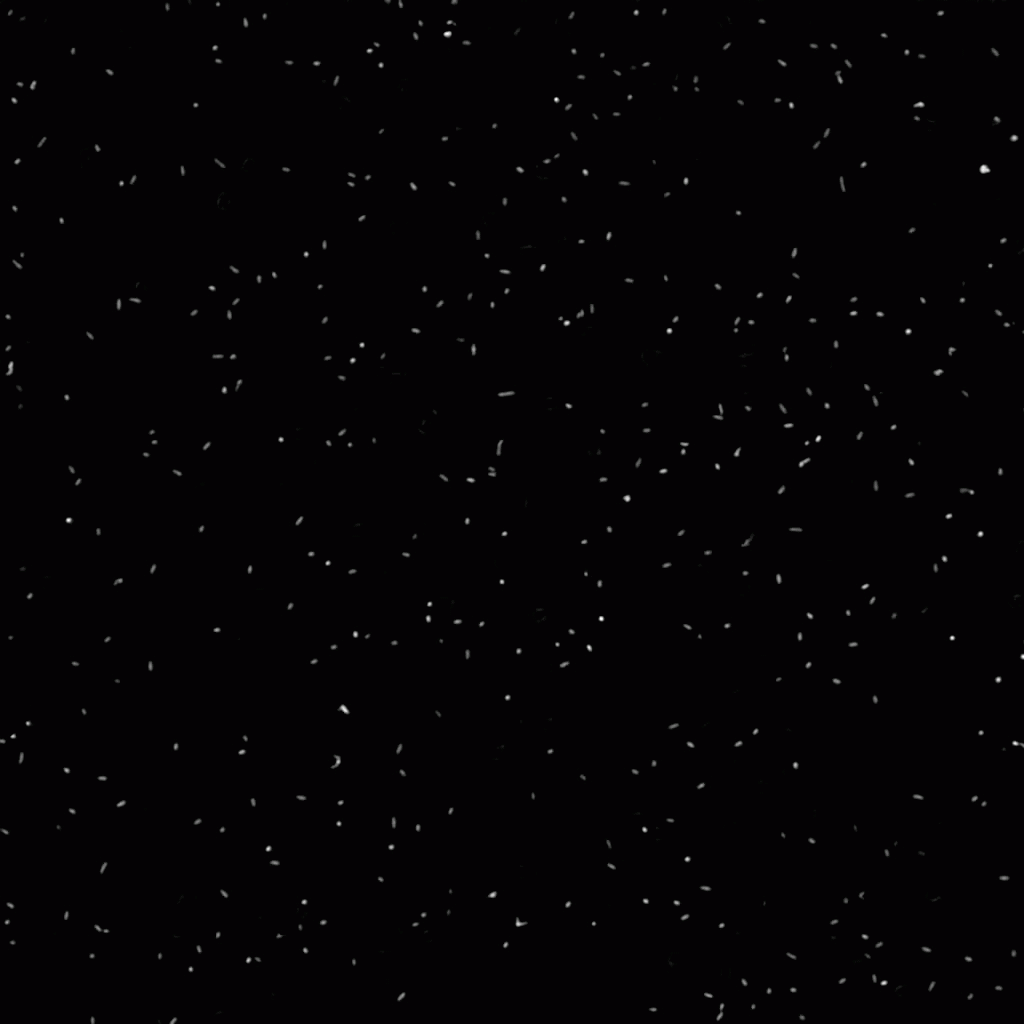
The position and orientation of rod-shaped bacterium can be determined, and each individual cell can be followed in time using tracking algorithms. An example is given below, with on the left the original image, and on the right the found position and orientation indicated in green. More information on algorithms to determine the positions and orientations of rods from microscopy images can be found here.
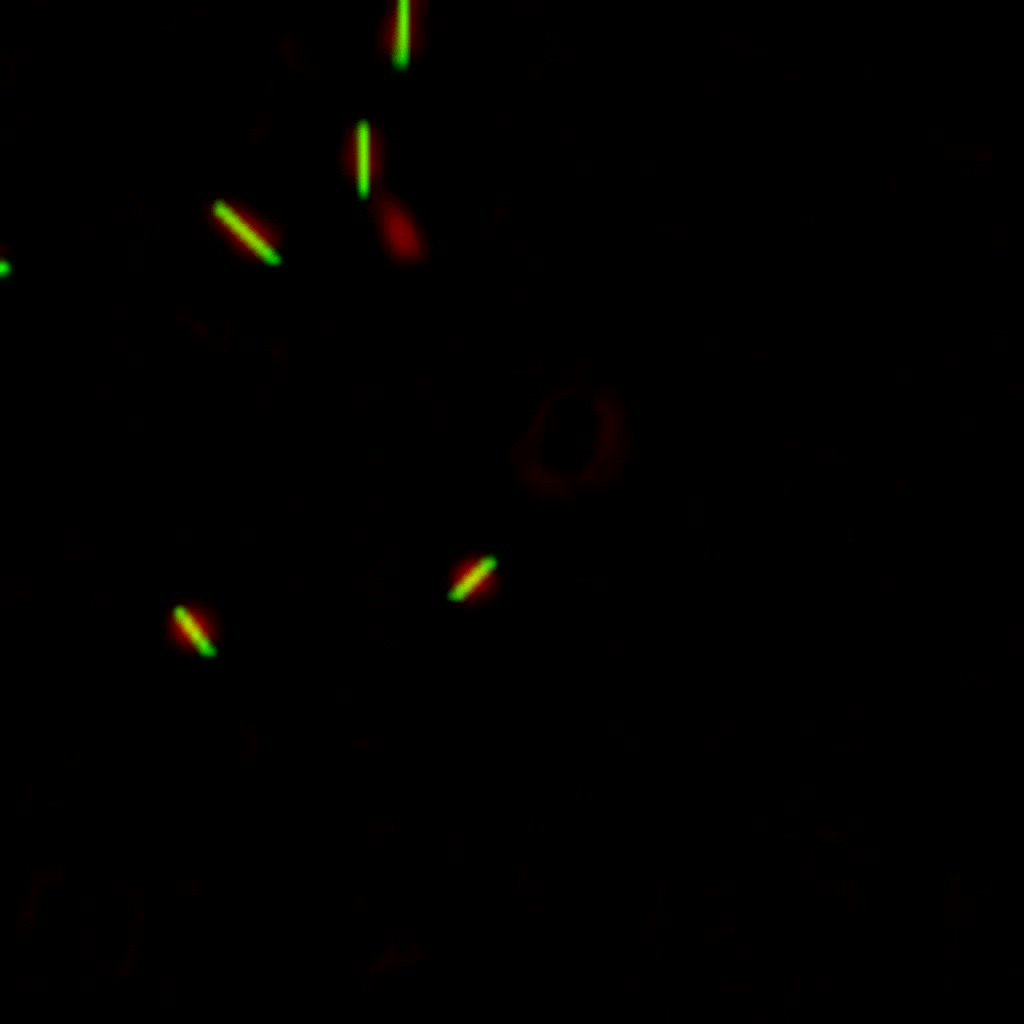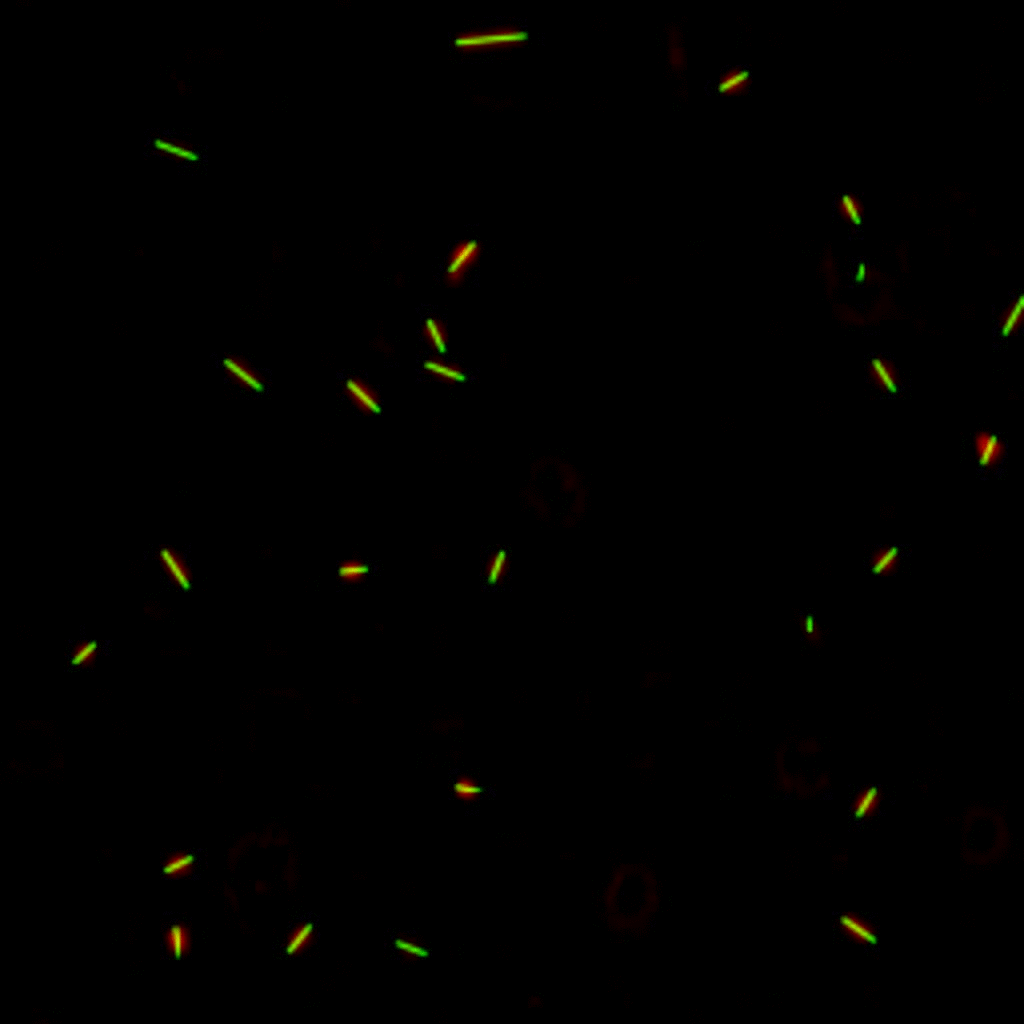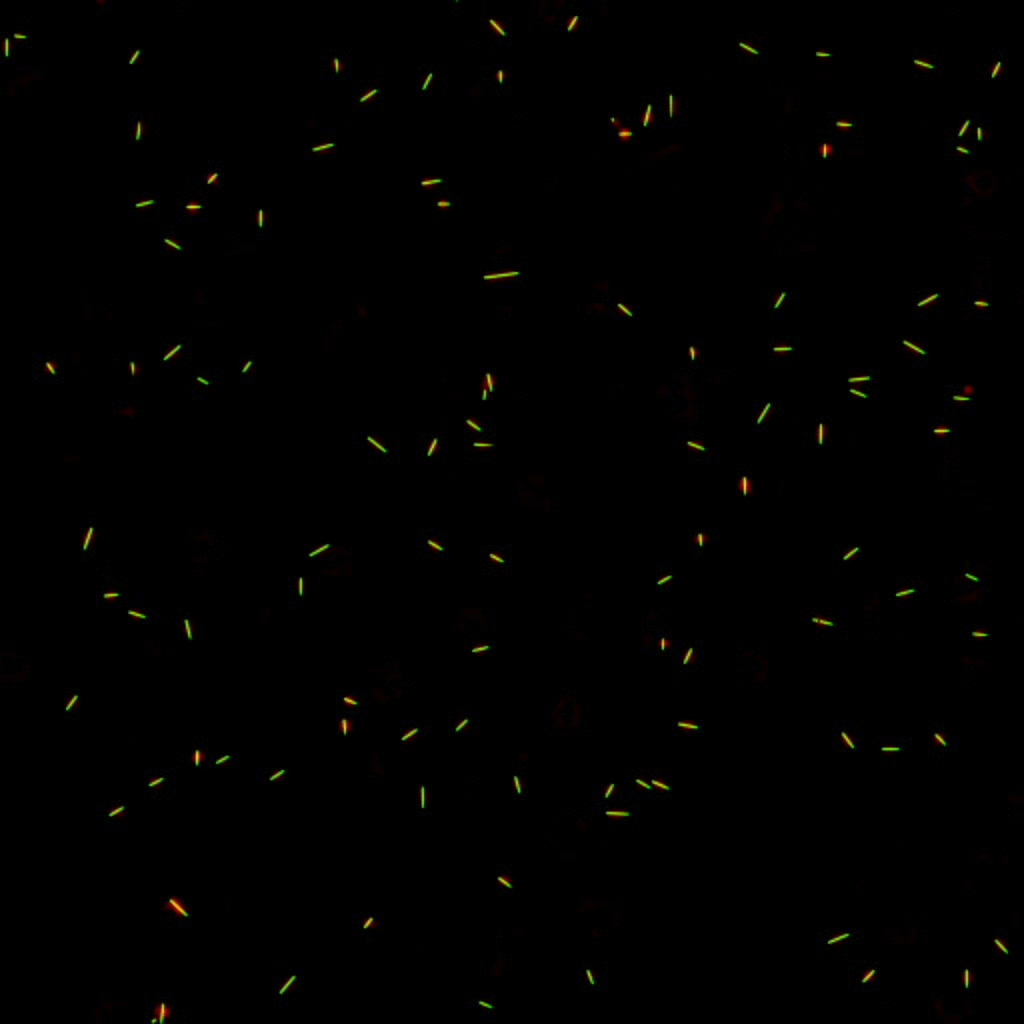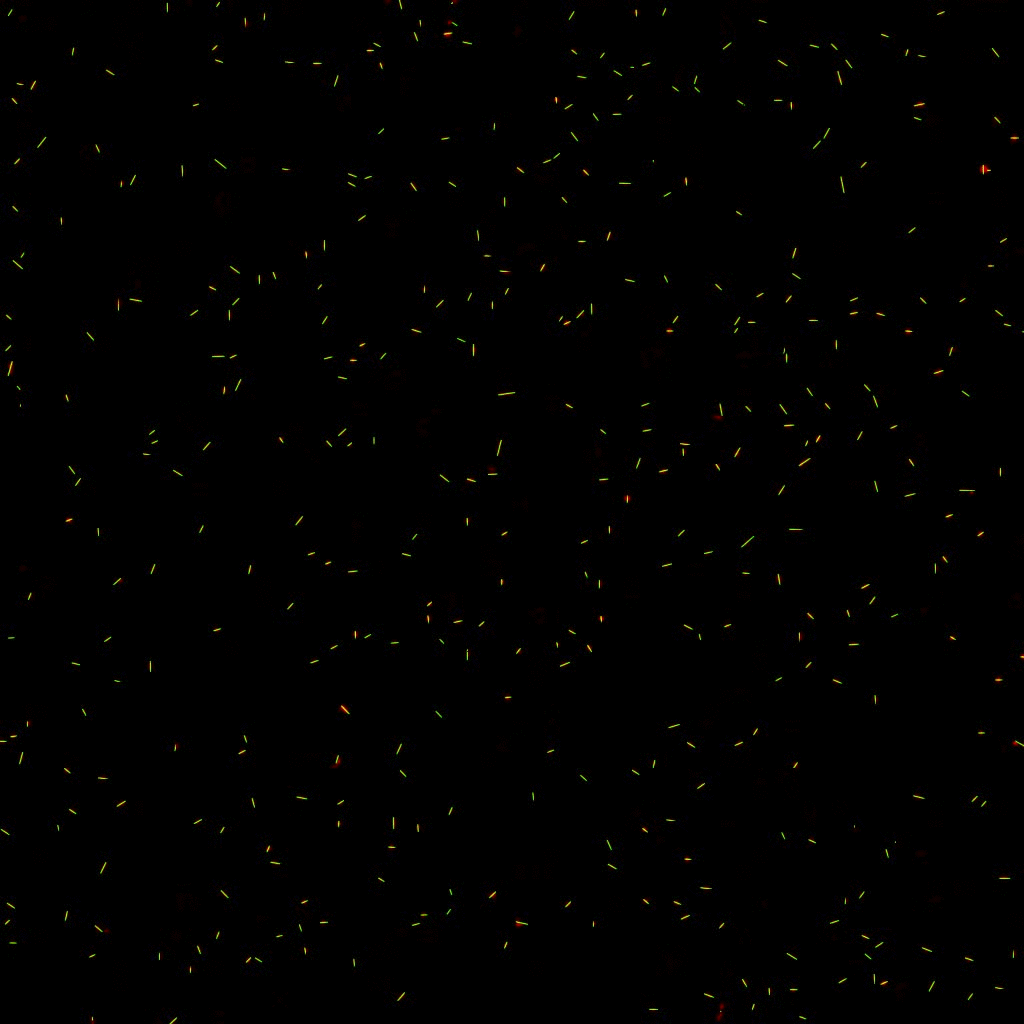
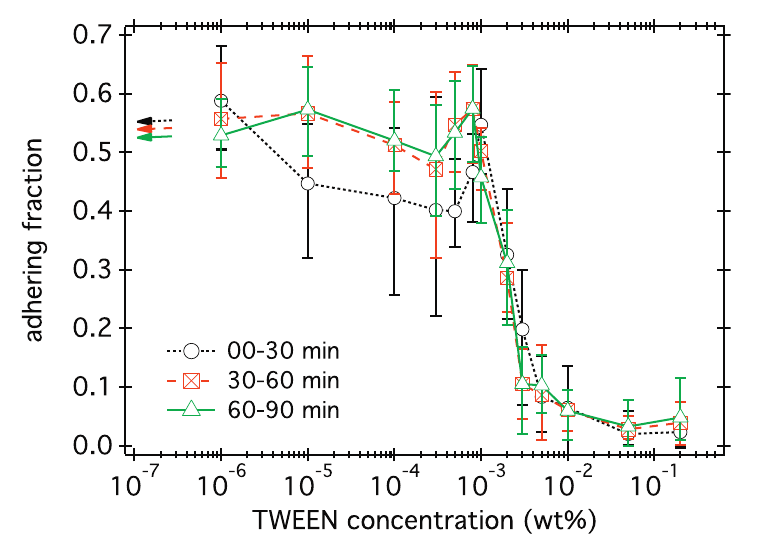
The adhesive interactions of bacteria with the surface are far from completely understood. In the case of E. coli above a glass surface, we have quantified how surface adhesion can be inhibited using TWEEN surfactant, a molecule with a hydrophilic head and hydrophobic tail. To do this, we used the extracted dynamics for each individual cell to automatically determine if it was adhering to the surface or not. The figure below (taken from a paper that can be found here) shows the fraction of bacteria on the surface that is adhering for different surfactant concentrations.
To look in more detail at the differences between individual cells, we have also used high-throughput methods to follow the surface interactions of tens of thousands of invididual cells in time above a glass surface. Although the bacteria in our experiments originate from a single clonal population (i.e. they are genetically identical), there are large differences between the adhesion dynamics of individual cells. Some of the cells in the experiment bind strongly to the surface whereas others adhere only weakly or not at all. The results can be explained using a patchy model where the adhesion strength depends on a number of adhesive patches that can be different for each bacterium. Non-adhering bacteria exhibit zero adhesive patches, weakly adhering ones bind through just a single patch, and strongly adhering cells can anchor themselves to the surface via one or more adhesive patches. The software that was used to identify and track rod-shapes bacteria in microscopy movies is described in detail in this paper.
Swimming bacteria
The surface of bacteria is much more complicated than that of most colloids. An important surface feature are flagella, thin filaments that can bundle together and propel the bacteria to swim. Flagella are typically several micrometers long but are to then to be seen in the phase contrast microscopy videos above.
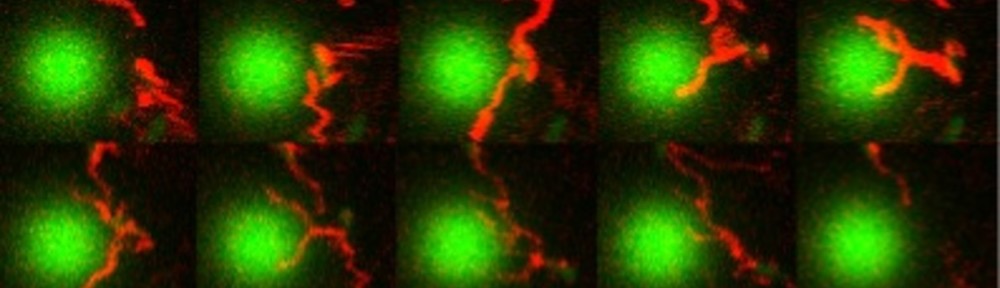
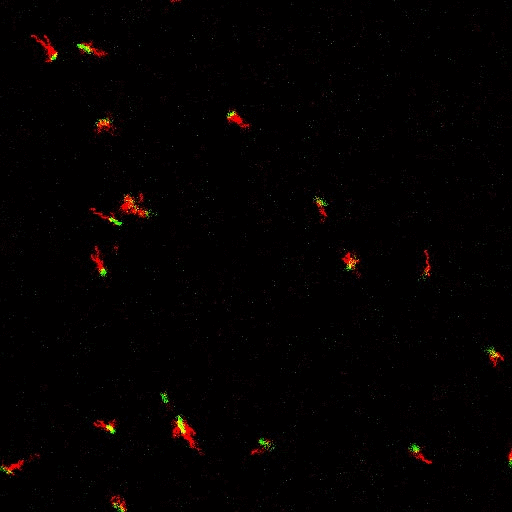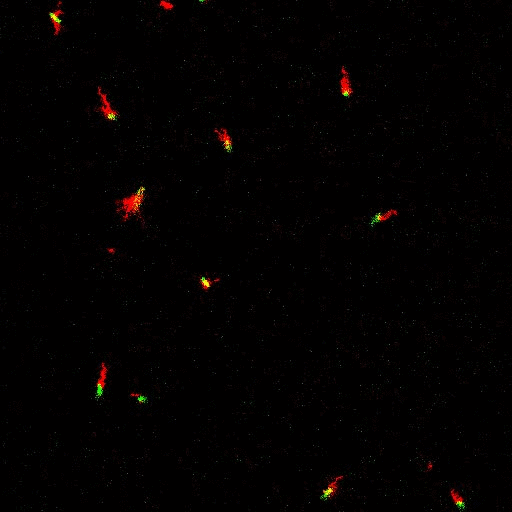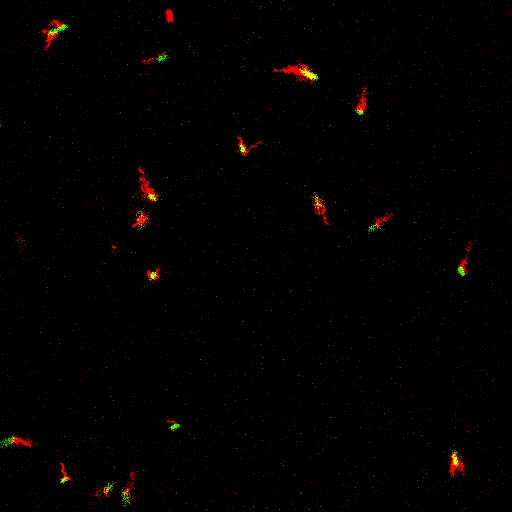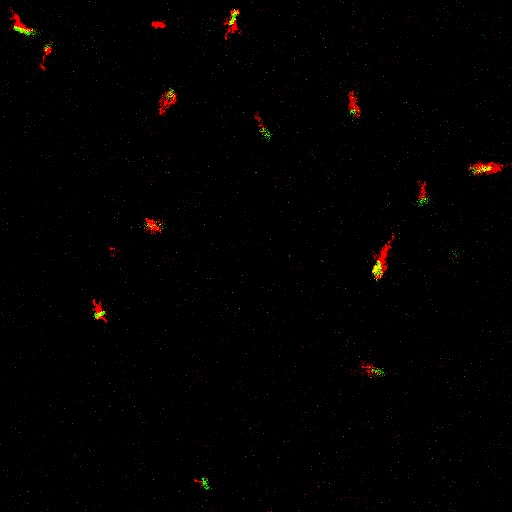
E. coli can be genetically manipulated to produce fluorescent proteins to stain their body, or to enable fluorescently staining of the flagella. The movie below shows a video of bacteria on the glass surface with eGFP staining (green) and fluorescently stained flagella (red). TWEEN surfactant is added to prevent surface adhesion).
Swimming bacteria on the surface describe circles, which is due to their flagella bundle rotating counter-clockwise and the body counter-rotating clockwise and pushing on the surface.
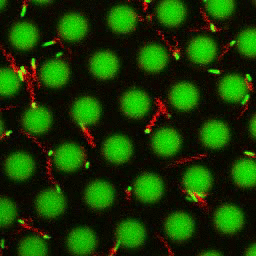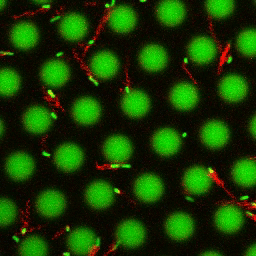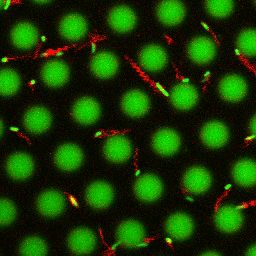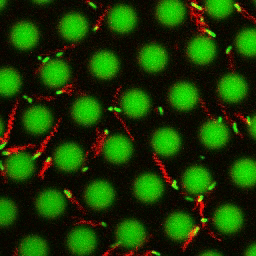
Bacteria swimming in a crystal
To see how swimming patterns are affected by obstacles, we performed an experiment where the bacteria are swimming just above a glass surface through a crystal of large 10 micrometer latex colloids (green spheres), click here for more details. The body of the bacteria is again shown in green and their flagella in red in the video below on the left. Although the distance between the colloids in the crystal is 10 micrometer, the colloidal spheres themselves appear smaller in the video because the centre of the spheres is not inside the focal plane. In the video below on the right, the trajectories of the bacteria by the tracking algorithm are indicated. The circular swimming motion of the bacteria is now inhibited by the large spheres that prevent them from turning to create more straightened trajectories.